Going Beyond Hit ID With Machine Learning & Del: The Prediction of Building Block Reactivity
Since X-Chem and Google’s seminal collaboration applying machine learning (ML) to DNA-encoded chemical library (DEL) screening output, ML has taken the industry by storm. The ability to drastically accelerate the discovery of campaign starting points with this approach has led many DEL operators to adopt ML+DEL as a hit ID strategy — in parallel to traditional DEL screen and compound resynthesis. At X-Chem, we’ve refined this pairing in our unique HITMiner offerings, which have proven track records of success in partner collaborations. These two offerings, however, are not the only ways we have integrated AI into X-Chem’s drug discovery pipeline.
Inclusivity and Fidelity: Our Uncompromising Approach
The success of any DEL or DEL+ML campaign can be traced back to library building blocks (BB) and the reactions that join them. Our library design philosophy has always been one of BB inclusivity, with the restriction that the synthetic fidelity of our libraries remains paramount.. Fortunately, using ML can expedite the choice of BBs for our library schemes without sacrificing inclusivity or fidelity!
X-Chem has a long-standing approach to ensuring the synthetic fidelity of our libraries:
- Experimental validation of each individual BB/reaction combination on a model DNA system
- Applying a yield filter for inclusion in library synthesis
This approach has resulted in 94% of our licensed compounds conforming to the fully elaborated library design.
Predicting BB Reactivity: Artemis AI in Action
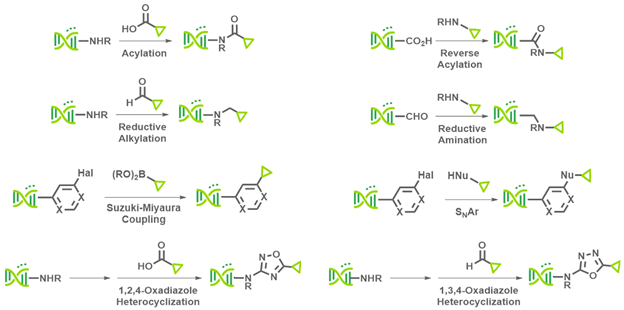
Since 2010, we have amassed tens of thousands of validation data points, and it eventually became apparent that this abundance of data would be particularly well-suited for predictions of BB reactivity using our proprietary AI engine, ArtemisAI. We conducted a retrospective analysis using data from eight “on-DNA” reactions (Figure 1) that spanned seven building block classes (fourteen experiments total). The results of this analysis revealed that X-Chem’s BB validation data and the predictive power of ArtemisAI pair very well.
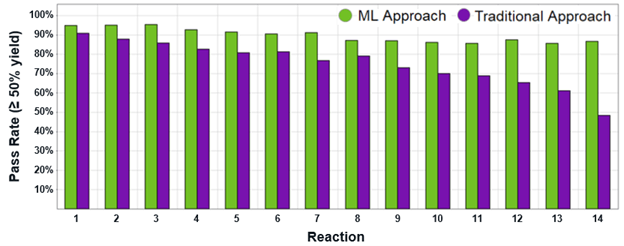
In the bar graph below (Figure 2), the “Traditional Approach” is the strategy of applying “brute-force” experimental validation to the entire set. The height of the purple bars (Y axis) indicates the percentage of BBs that “pass” (have yields in excess of 50% for that reaction). In the “ML approach,” we train a suite of ML models within ArtemisAI on a portion of the data, then predict which of the remaining BBs will pass. The height of the green bars indicates the percentage of those predicted passes that are experimentally confirmed.
We observed that the ML approach consistently identifies passing BBs at a higher rate than the traditional approach — for all 14 reactions.
We recognize that BBs are the key drivers of diversity in our libraries, and we are devoted to constantly expanding our collection. Attrition is expected in any large-scale acquisition of BBs because some fraction of BBs will fail individual validations. These BBs are therefore not included in future libraries utilizing this chemistry. However, using our ML approach, we can identify BBs within the candidate set that are likely to pass validation prior to investing in their acquisition. This enables two critical benefits:
- The enrichment of each set in high-yielding BBs
- Continuous expansion of the diversity of our libraries
Library Diversity: An X-Chem Priority
The synthetic reactions we use to connect our BBs confer structural diversity to our library schemes. We even prioritize the development of reactions that employ numerically large BB classes because they offer the most value in the form of diversity. However, the experimental validation of tens of thousands of amines for a new cross-coupling, for example, can be an arduous task for even the most efficient library chemist.
: Certain reactions can be fickle (try handing a chemist a list of nucleophiles and asking them to predict their competency in an SNAr reaction!). The only way to ensure that 100% of the BBs that pass validation are included in the libraries (and no others) is to take the brute-force approach. However, if we loosen that threshold slightly, the advantages of ML and experimental validation working in tandem become significant. That’s why we designed a process that can tolerate even the most unpredictable reactions.
ML and Experimental Validation: Iteratively Reducing Timelines
We use an iterative active learning loop, consisting of (a) alternating rounds of experimental validation to generate data and (b) ML model generation and predictions of validation outcomes. Then, we experimentally validate the predicted minority class, allowing us to bolster the training data with data points that the model is least confident in predicting. We then continue with ML model generation and prediction again, and so on.
This process can be iterated using the ML evaluation metrics to accept predictions on the remaining — untested — BBs. This approach translates to weeks of library development time saved!
BB Reactivity: One Application Among Many
Predicting BB reactivity is just one of the ways that X-Chem is applying ML to exponentially accelerate our delivery of drug discovery services. We are constantly expanding our AI/ML capabilities beyond hit ID, and this commitment has already impacted numerous areas of our business. Stay tuned for more exciting advancements!
2024: A Year of Scientific Excellence for X‑Chem
One of the best parts of being part of X-Chem is seeing all the great science being conducted around the...
The Importance of Selecting the Right CRO Partners for Drug Discovery Success
When it comes to drug discovery and development, having the right team and partners is critical for success. For a...